Do you ever feel like the age of genomics has made dairy cattle breeding more complicated? Or perhaps more scientific? While genomics brought us accelerated progress, it has also introduced us to a range of terms previously used by scientists alone.
“Haplotype” has become a household name used in bull catalogs and communication by evaluation centers. But it is probably one of the most misunderstood scientific terms in the industry. Often used in the same breath as “recessive,” “genetic defect” or “genetic condition,” haplotypes have gotten a bad reputation, which is not completely fair.
To correctly use this new information in genetic herd management, it’s good to explore what haplotypes are, what they are used for and if they really are all bad.
What is a haplotype?
The definition of a haplotype is very simple, and it has little to do with genetic defects. DNA consists of base pairs. A haplotype is simply a short section of DNA, or a sequence of those base pairs, passed on to the next generation. If you would cut up the strand of DNA into bits, each bit would be a haplotype if it is more than one base pair and the whole segment is inherited from either the mother or father (Figure 1).
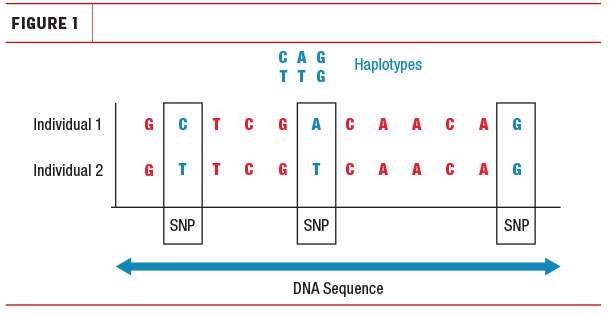
In genomic selection, we do not read individual base pairs; instead we use genetic markers – most often single-nucleotide polymorphisms (SNPs) scattered across the genome. Therefore, when we refer to a haplotype for a genomically tested animal, we are talking about a sequence of SNPs, which signifies a piece of DNA inherited from either sire or dam.
Therefore, a haplotype is not a gene or even part of a gene. A haplotype can contain one or multiple genes – or none at all.
Are all haplotypes genetic defects?
No, a haplotype can contain a positive or negative genetic variant.
Our industry has inadvertently made the word “haplotype” synonymous to genetic defect, while it actually isn’t. This has given the word “haplotype” its negative reputation.
We use haplotypes as a handy tool to trace genes or genetic variants that cause the expression of a particular phenotype. In some cases, this is a desired trait, and in other cases it is detrimental or lethal.
Because genetic defects have such a large impact, they are more often discussed than desirable traits and, thereby, haplotypes are most often related to genetic defects. But haplotypes are also used to identify desirable traits such as polled and coat color.
So why do we talk about haplotypes and not genes?
There are multiple answers to this question, but one of the main reasons is: We simply do not know what genetic variant is causing the phenotype.
Our commercial genotypes mainly consist of SNPs, which means that unless the causal genetic variant is a specific SNP we have mapped, we do not know what causes the observed phenotype. When we have enough animals with the distinct phenotype, we can track where on the genome the variant likely is and dial in to a particular part of related DNA – the haplotype. We then know that this haplotype likely contains the causal variant, but we do not know what it looks like without genome sequencing and additional research.
This is often the reason why a genetic variant is first presented as a haplotype. Testing for the particular haplotype allows breeders to get a good sense of whether their animal may be a carrier for the new genetic variant.
If possible, researchers then sequence the DNA of affected animals and try to find the causal gene. When found, it is then a matter of getting the variant on the DNA chips used for genotyping, which can be a complicated process.
However, even when the genetic variant is known and a gene test is available, haplotype tests often remain offered because they present a much lower-cost way to test for carrier status. Also, if we were to add gene tests as soon as they were discovered by researchers, that would require frequent updates to commercial genotyping tests, which is not practical.
Why are haplotype tests not 100% accurate?
Especially for new genetic variants, haplotype tests are not 100% accurate.
Not all animals are genotyped, and not all animals are genotyped using high-density SNP chips. We use mathematics and pedigree data to predict and fill in SNP markers that we are missing in our haplotype of interest. Our haplotypes and the result of a haplotype test thereby depend partly on the quality of pedigree data and how many genotyped animals exist in that pedigree data. Data is added continuously, which means that a haplotype result can change when there is pedigree added or parentage was corrected in the lineage of the specific individual. In addition, new genetic variants may not have a large number of known carriers, which means the identified haplotype may be relatively long and thereby less exactly pinpointing the causal variant. As more carriers are discovered, we can dial in the exact coordinates of the haplotype and make our prediction more accurate.
Ideally, we would test every genetic variant with a gene test. This would remove much of the insecurity and frustration breeders experience. That said, this would also be a lot more costly for breeders. The haplotype test allows us to detect carriers fast and at almost no cost at a reasonable reliability level of 95% to 99%. As more animals are genomic tested and DNA arrays improve, reliability levels of haplotypes and the potential to detect genetic variants improves as well.
The slightly lower reliability level of haplotype tests is also the reason some breed associations choose not to place haplotype results on any official export documents. And it is the reason why it is possible that a haplotype test result differs from the gene test result when a gene test can be performed.
What is a lethal haplotype?
A lethal haplotype is most often a lethal recessive we are testing using a haplotype.
The HH1 to HH6 haplotypes in Holstein, JH1 in Jerseys and AH1 are examples of lethal recessive haplotypes. These genetic defects cause the embryo to die when two carriers are mated. Most often, this happens so early in gestation it simply seems that the animal has come back in heat. We therefore say these lethal recessive haplotypes affect fertility.
Are all lethal haplotypes equally bad?
All detected lethal recessive haplotypes have a proven effect on fertility. However, this effect is not equally bad, and the chance of this effect occurring is not equally high. The chance of a carrier mating depends on the frequency of the gene in the population. Each of the published lethal recessive haplotypes have a different allele frequency. For example, 1.92% of U.S. animals carry a copy of HH1. This frequency is 2.22% for HH5 and 0.37% for HH4. All lethal haplotype frequencies are published by the USDA and can be found online.
Should I stop using bulls with a carrier status for lethal haplotypes or recessives?
No, not necessarily. The chance of carrier matings is low for the majority of lethal haplotypes. To continue with the example of HH4, only 0.37% of U.S. Holstein animals carry a copy for HH4, and those animals have to be mated with each other for HH4 to be expressed. With the availability of genomic testing and a multitude of mating programs, carrier matings can easily be avoided.
Any genetic defects with large effect and high frequency get quickly taken care of by breeding programs from A.I. providers. Bulls are tested at a very young age and culled when carrying detrimental genetic defects. Pretty much all U.S. A.I. companies publish carrier statuses on every bull page for your information.
Unfortunately, not all A.I. companies worldwide publish carrier statuses of their bulls, and there is no international standard with how carrier statuses should be expressed either. This includes the denotation whether the result comes from a haplotype or gene test. When you are missing information or information is not clear, ask your A.I. provider how and for what the bull was tested.
The bottom line
Scientific jargon has entered our industry at a rate that makes it difficult to keep up on what everything means. When it comes to managing your herd, don’t be too scared of “haplotypes,” as their bad reputation is not entirely justified. Know that not all haplotypes are bad, and not all bad haplotypes are equally bad.
In addition, feel safe in knowing there are measures in place to detect any new detrimental genetic defects. Data on genetic variants is publicly available, and mating programs allow for easy avoidance of carrier matings of genetic defects. Also, feel free to ask if a genetic variant was tested using a haplotype test or gene test. When it comes to genetic management, it’s always good to know what exactly you are looking at and how it can be managed to get the best result for your herd.






