To read this in French, click here.
A healthier herd. It’s a simple-sounding concept that’s proven difficult in practice.
Finding genetics that resist routine pathogens across environments on dairy farms has been a focus of the industry over the last 10 years. In that time, there has been a shift from broad health traits based on culling and death data toward much more specific traits for individual diseases. Alongside this has been the development of immune response traits, examining the direct biological ability of an animal to react and respond to a disease challenge at the cellular level.
The result is, we now have multiple ways to evaluate and potentially solve the big question. Currently, multiple genetic evaluations are carried out in parallel with the same goal – to identify the genetics that will reduce herd disease.
One of the greatest challenges in improving health from genetics is collecting high-quality data and enough of it to accurately reflect genetic variation in the population. Health traits are hugely influenced by environmental factors and are not consistently recorded from herd to herd. This creates a great challenge in getting one good genetic evaluation that encompasses a disease, or many disease phenotypes, on-farm.
Selecting for health traits
Data was recently collected from a group of 100 large commercial farms around the world – all with high-quality and consistent disease records on their animals. In total, the dataset represented 330,000 cows and over 160,000 excellent health records.
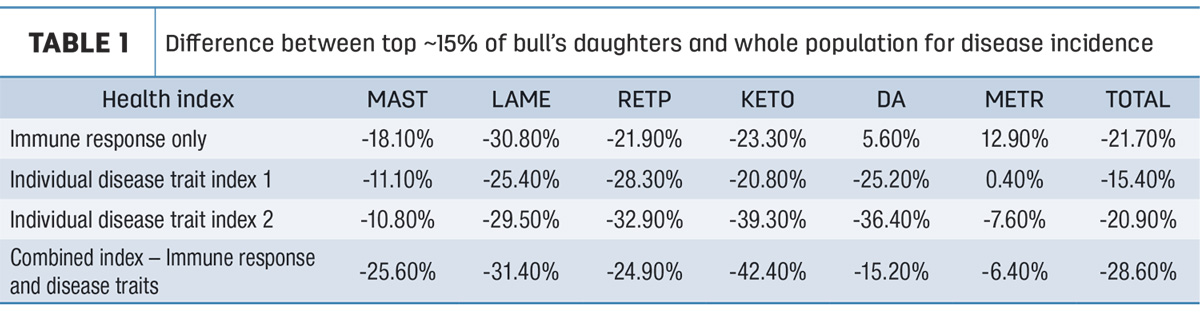
The study analyzed the performance of daughters of sires based on the sire’s health indexes (based on individual disease traits), and their genetic evaluations for immune response (a combination of antibody-mediated, cell-mediated and innate immune system components). Results were highly variable by herd and by disease. In general, direct immune response outperformed individual disease traits when analyzing total cases of recorded disease in the population. Some traits, especially those with metabolic basis (ketosis, displaced abomasum) favored selection on individual health traits. Overall, direct immune response performed best, but for a given herd, there wasn’t a clear answer of which genetics are always best.
Can a combination of health genetics outperform individual indexes?
To combine multiple traits for one goal, one would usually utilize the genetic correlations between traits to find a combination that maximized genetic progress for the trait in question. In this case, disease resistance, or, more aptly, disease cost reduction.
For this dataset, however, it wasn’t possible to calculate genetic correlations between different disease traits and immunity. Instead, big data was used to optimally combine these traits into an index best suited to minimize disease costs across all farms. An algorithm (a machine learning/artificial intelligence technique) was used to find the ideal combination of traits to minimize the expected disease cost per animal in the population. The higher the index value, the lower the expected number of cases and total cost of disease.
Results found that the combined index outperformed each individual component on its own by a significant margin. Comparing the top approximate 15% of bulls for each genetic measure to the rest of the population, the combined index had 28.6% less disease in total, compared to 21.7% for immune response alone, and 15% to 20% for individual health trait indexes. Full results can be seen in Table 1. Essentially, the top 15% were more resistant to disease when selected based on the combined index.
Why does it work?
You may ask why the combination of these traits is able to outperform any of them on their own. For example, health traits were designed to predict this exact type of data – isn’t that enough?
The core advantage of combining genetic evaluations into one comes from the complexity of the immune system itself. To aptly fight off a pathogen, the immune system needs to be able to do two core things:
- Mount a strong immune response, meaning they send a lot of immune defense molecules to the site of infection that can kill off the invading pathogen (immune response)
- Recognize the invading pathogen and be able to know that the cell itself, in the case of a bacteria, or the contents of a cell, in the case of a virus, are harmful and should be dealt with (individual disease traits).
This means that selection for a combined index will lead to a healthier herd faster, translating to less treatment costs, less lost milk and longer-lasting cows. Estimates from initial data have shown that for each standard deviation improvement in a combined immunity index, a cow is expected to save over $160 over its lifetime from reduced disease. Studies have also shown a greater disease reduction, and a greater economic impact from a combined index, rather than any individual set of health traits.
No health trait is perfect, and each one has its own benefits and drawbacks depending on the size, scope and quality of the data that go into a genetic evaluation. It is clear, however, that the combination of these parts is greater than the individual components.
Combining evaluations will lead us more quickly to a healthier herd.







